Programmable Cytometry
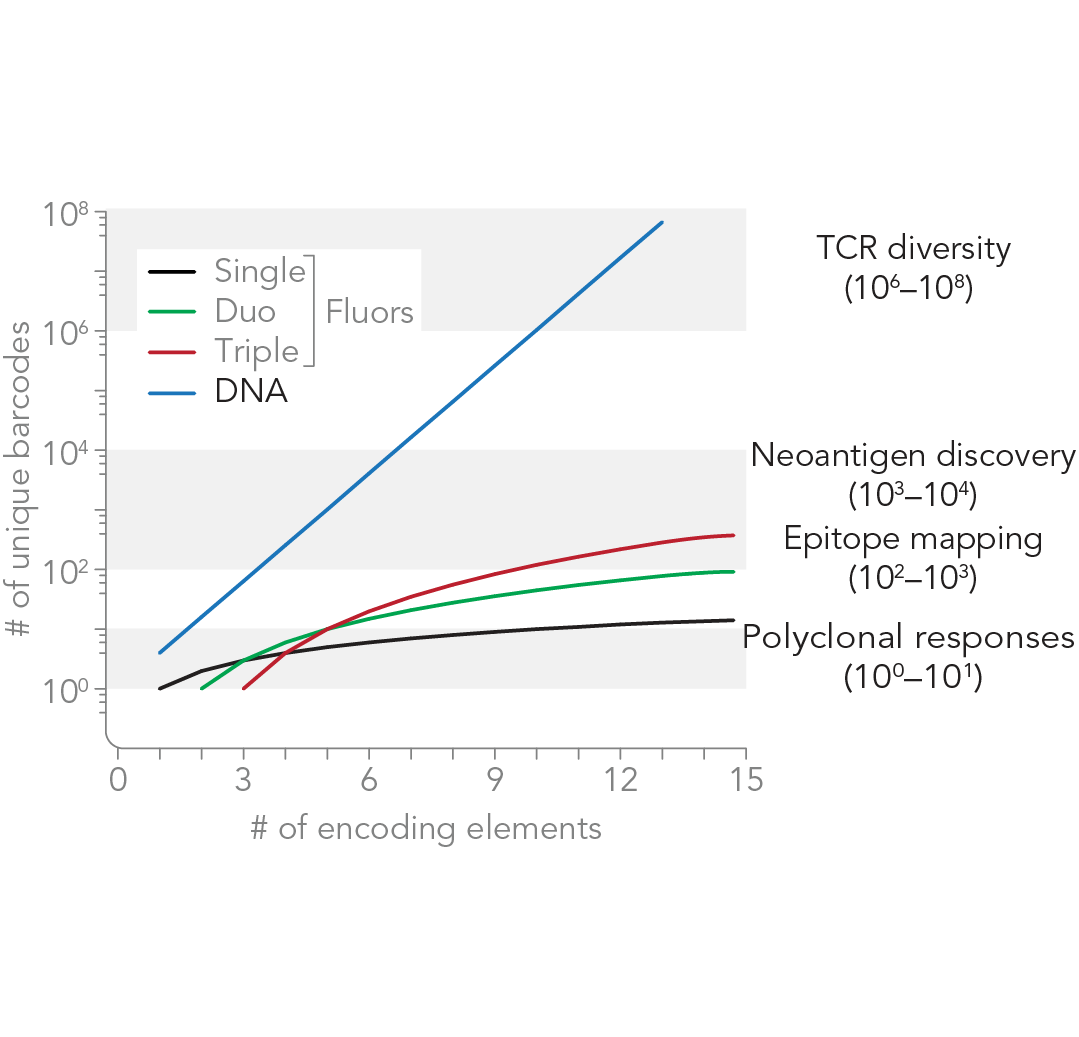
Single cell cytometry has fundamentally increased our understanding of immunobiology and led to myriad biomedical applications. However, analysis of the immune repertoire remains challenging as T cells alone comprise an estimated diversity of one to one hundred million different antigen-specific T cells at frequencies as rare as one in one million.
To address these issues, we are developing programmable cytometry, a strategy where DNA-based constructs are used for cell analysis in contrast to optical labels. This approach provides the ability to construct massively multiplexed libraries at a scale that exceeds that of optical labels by orders of magnitude.
Our studies show that this approach is sensitive to single antigen-specific T cells [1] and allows nondestructive sorting of immune cells [2]. We are applying these technologies to study immune responses to acute and chronic viral infections such as HIV and cancer immunotherapies.
[1] “DNA-barcoded pMHC tetramers for detection of single antigen-specific T cells by digital PCR” Anal. Chem. 91(4), 2695–2700 (2019).
[2] “Individually addressable and dynamic DNA gates for multiplexed cell sorting” Proc. Natl. Acad. Sci. USA 115(17), 4357–4362 (2018).
Protease Biocircuits
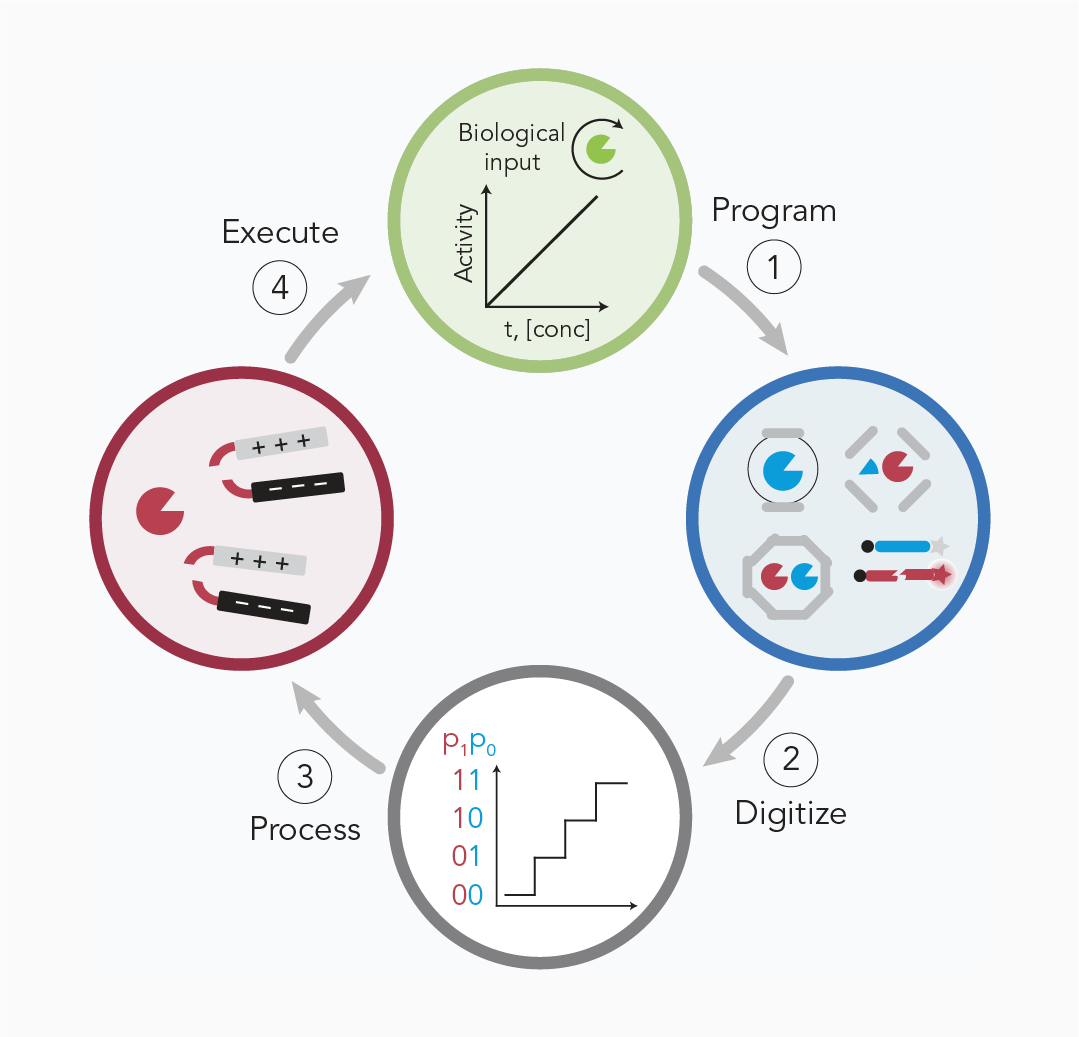
Biological circuits engineered to interface with living systems as plug-and-play constructs will enable new applications for programmable immune therapies and diagnostics.
We are exploring how biomaterials that treat proteases as biological bits could be used to design biocircuits for medical applications. We recently demonstrated biomaterials that function as analogs of electronic components such as logic gates and analog-to-digital converters for use in drug delivery or disease detection [3].
We are extending these ideas to develop antimicrobial strategies that rely on protease-activatable prodrugs. By modeling bacterial treatment success and failure, we are designing logic-based treatment strategies to overcome resistance [4].
[3] “Protease circuits for processing biological information” Nat. Commun. 11, 5021 (2020).
[4] “Dimensionless parameter predicts bacterial prodrug success” Mol. Syst. Biol. 18: e10496 (2022).